DNPLab is an object-oriented Open Source Python-based package for importing, processing, and analyzing data determined in a Dynamic Nuclear Polarization (DNP-NMR) experiment. The aim of the project is to provide a free, turn-key python-based processing package DNP-NMR data. For more information visit:
- the GitHub repository,
- the Online Documentation,
- or follow us on Twitter for updates.

To compensate for magnetic field drifts a lock-channel is commonly used in high-field NMR spectrometers. This is standard practice in solution-state NMR spectroscopy, but has also become more popular in solid-state NMR spectroscopy to achieve high-resolution NMR spectra.
In low-field NMR spectroscopy, the technique is less common. In ODNP spectroscopy in particular, often a single-channel NMR spectrometer is used. Furthermore, these experiments are often performed in electromagnets or permanent magnets and magnetic field drifts can be a challenge to achieve high resolution 1D or 2D spectra.
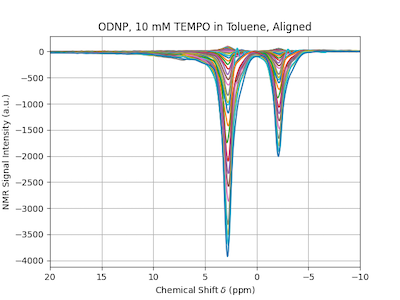
DNPLab has an integrated feature to align spectra. Spectra can be aligned in the frequency domain using a Fourier Transform Cross Correlation function. The following example demonstrates this.
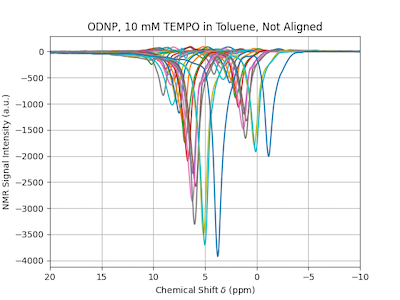
Shown in is a series of ODNP-enhanced 1H-NMR spectra of 10 mM TEMPO in Toluene. Two peaks are visible, one for the methyl group protons and one for all the aromatic protons. Due to the magnetic field drift, the spectra are not aligned. In this case, automatic analysis of the data is not possible.
With just a single line of code the spectra can be aligned using the DNPLab built-in function align.
data_aligned = dnp.align(data)
That’s it. For more details check out the DNPLab example on how to align spectral data. For more processing tips and tricks check out the DNPLab Documentation or follow us on Twitter for updates.
