DNPLab is an object-oriented Open Source Python-based package for importing, processing, and analyzing data acquired in a magnetic resonance experiments. Typical experiments include Electron Paramagnetic Resonance (EPR) spectroscopy, Dynamic Nuclear Polarization (DNP) spectroscopy, and Nuclear Magnetic Resonance (NMR) spectroscopy.
The aim of the project is to provide a free, turn-key python-based processing package for DNP-NMR data.
DNPLab is created by Bridge12 in collaboration with the Franck Lab (Syracuse University) and the Han Lab (NWU).
If you like to stay updated, follow us on Twitter or head over to the GitHub repository.

Why DNPLab?
The field of DNP-NMR spectroscopy is relatively new and instrumentation can vary largely from commercially available DNP-NMR systems (e.g. gyrotron-based DNP-NMR spectrometers) to home-built instruments. Experimental data is often in different formats (e.g. TopSpin, VnmrJ, Delta, Prospa, …) requiring different import tools and additional information is often stored in separate log files (e.g. temperature, or microwave power). DNPLab makes importing, processing and analyzing DNP-NMR data simple. Import data from any major NMR platform, use the built-in functions for processing, or add your own code.
DNPLab is Open-Source
DNPLab does not rely on proprietary software and is entirely written in Python and can easily be installed by running the command pip install dnplab in a terminal. The package is entirely open source and the source code is available for download from GitHub. Updates are released on a two-week cycle and the Online Documenation is constantly updated.
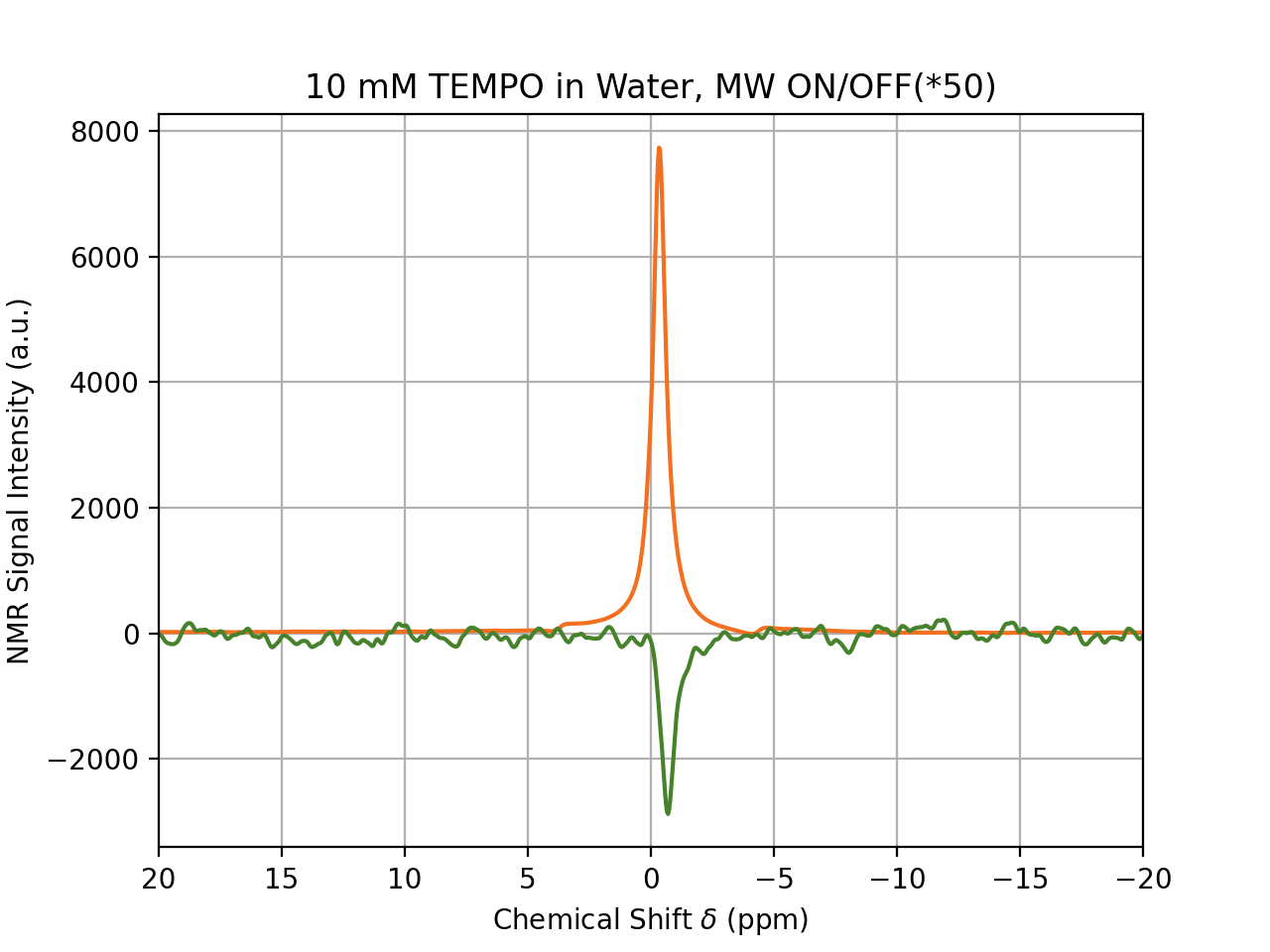
Analyze DNP-NMR Data …
DNPLab is very simple to use. With just a few lines of code, data can be loaded, processed and plotted in a publication-quality figure.
import dnplab as dnp
data = dnp.load("data.1d")
data = dnp.remove_background(data)
data = dnp.apodize(data, lw=15)
data = dnp.fourier_transform(data)
dnp.fancy_plot(data, xlim=[-30, 30])
You don’t have to be a Python wizard to use DNPLab to process your data … if you are, we still think DNPLab will be very useful to you. The complete example can be found in the Online Documentation.
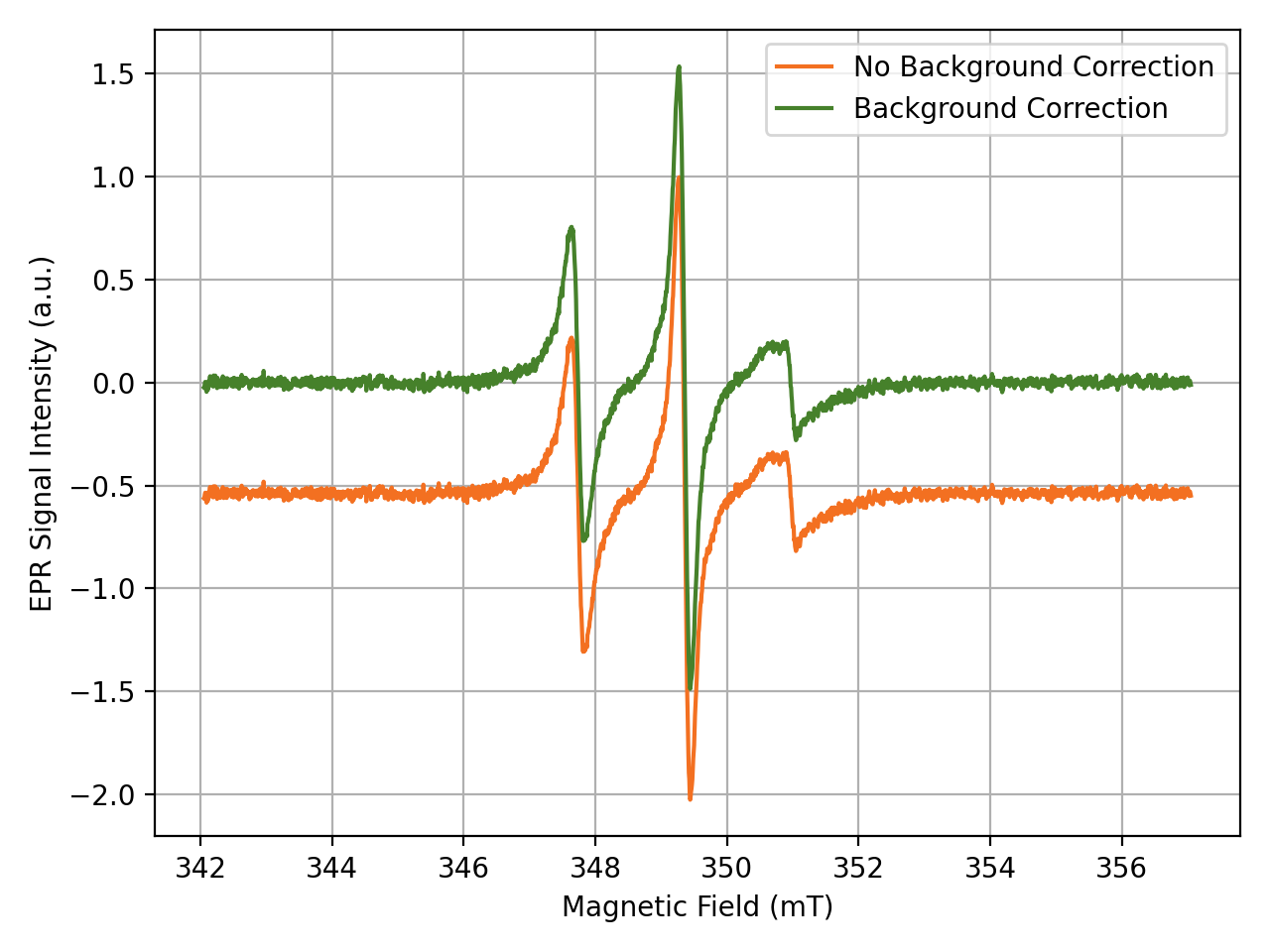
… and Plot EPR Spectra
To understand the DNP process, it is crucial to understand the EPR properties of the polarizing agent. With DNPLab you can easily import EPR spectra from any commercial or home-built spectrometer and no additional software is required. For more features check out the Online Documentation.

Get Involved
DNPLab was created by Bridge12 scientists in collaboration with scientists from the Franck Lab (Syracuse University) and the Han Lab (UCSB). It is currently maintained by a small group of people. However, we are constantly looking for volunteers to help out. If you like to get involved feel free to reach out to info@bridge12.com.
Technical Specifications
- Easy to install using pip:
pip install dnplab - Open source code available on GitHub
- Documentation available online